Parametric NAFEMS LE10 benchmark
Comparison of resource consumption for different FEA programs
1 Introduction
This test aims at comparing CPU and memory consumption when solving the same problem with different cloud-friendly finite-element analysis programs. Particularly, it was designed to understand how FeenoX compares to other well-established tools and to understand where and how to optimize the code.
The programs tested are
The problem being solved is the NAFEMS LE10 problem. It was chosen because
- It is a well-established benchmark since its publication in 1990
- It is simple yet has displacement boundary condition on an edge in addition to faces that makes it challenging
- The reference solution is a single scalar which is easy to compare among different approaches
Each program solves the problem parametrically over a wide range of mesh refinements using two types of Gmsh-generated second-order grids:
- locally-refined (around point D) unstructured curved tetrahedral grid, and
- straight incomplete (i.e. hex20) fully-structured hexahedral mesh.
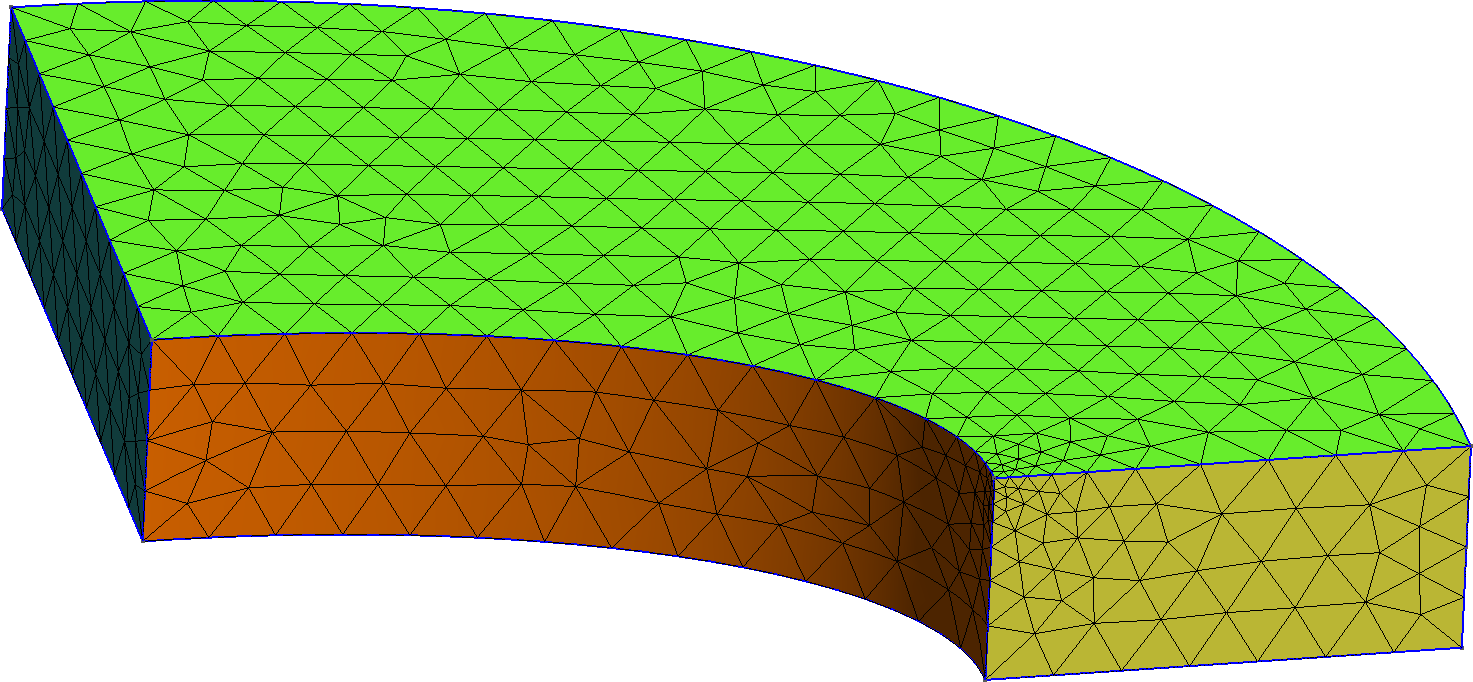
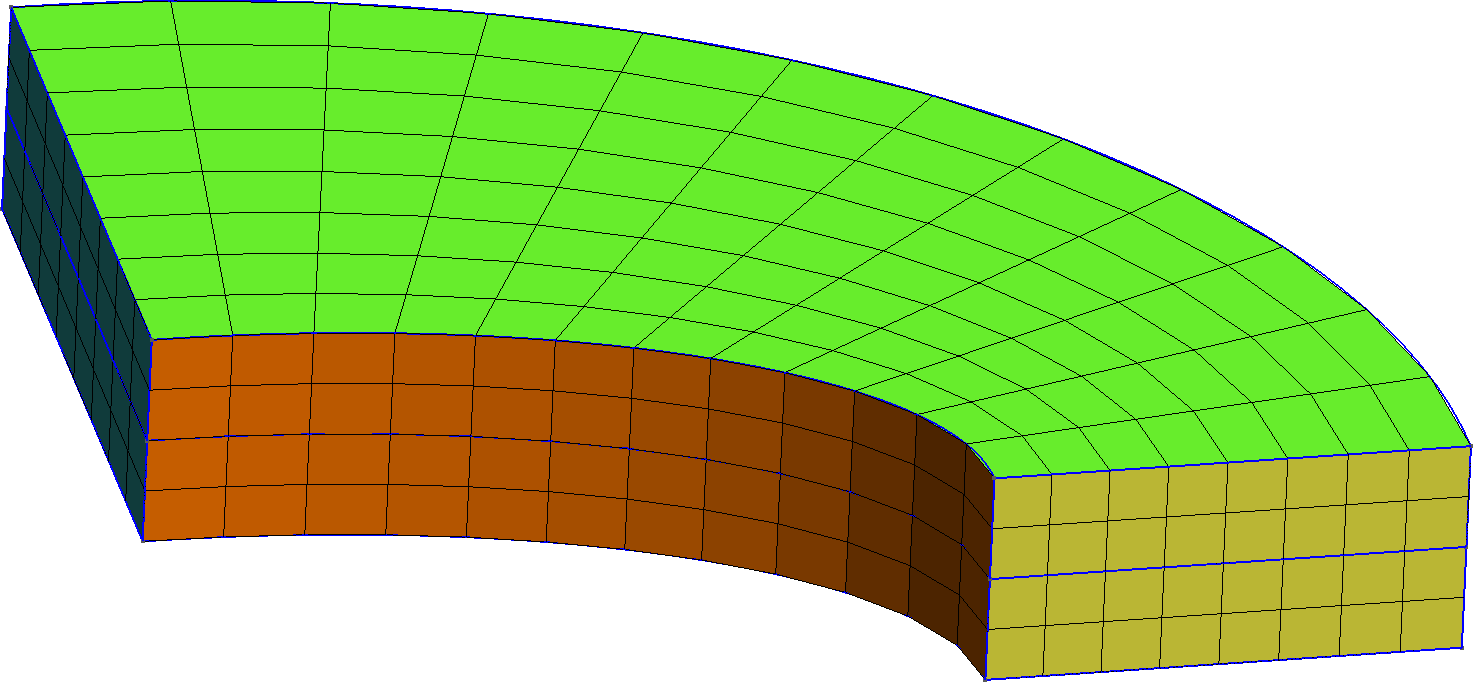
Figure 1: The two types of meshes used in this test. a — Tetrahedral mesh, b — Hexahedral mesh
The NAFEMS LE10 problem asks to compute the normal stress in the y direction at point D that has coordinates (2000~\text{mm},0,300~\text{mm}). For each mesh type (tet/hex), a refinement factor c \in (c_\text{min}:1] is applied. Besides \sigma_y, the wall time, CPU time and memory are recorded for each run so as to create plots of these results vs. c and vs. the total number of degrees of freedom being solved for—which can be different in each code even for the same c, as explained below.
This way of executing FEA programs follows the FeenoX design basis of being cloud-first (and only later desktop-friendly). It is mandatory to be able to control the execution and read the output from an automated script. The reasons for this requirement are explained in the FeenoX documentation, particularly in the SRS and SDS. It might happen that some of the codes tests seem to need to setup and/or read the results in a unnecessarily complex and/or cumbersome way because they were not designed to be either cloud-first and/or script-friendly. It might also happen that the cumbersomeness comes from my lack of expertise about how to properly use the code.
Even though there are some particular comments for each of the code used in this comparison, this test is not about the differences (and eventually pros and cons) each code has for defining and solving a FEA problem. It is about comparing the consumption of computational resources needed to solve the same problem (or almost) in the cloud. The differences about how to set up the problem and considerations about usage, cloud friendliness and scriptability are addressed in a separate directory regarding benchmark NAFEMS LE11, that involves defining a temperature distribution given by an algebraic expression (under preparation at the time of this writing).
1.1 Reference solution
The original problem formulation (which can be found in one
of FeenoX’ annotated examples) states that the reference solution is
-5.38 MPa. This can be confirmed with FeenoX using the input
le10-ref.fee.
$ gmsh -3 le10-ref.geo
[...]
Info : Done meshing order 2 (Wall 0.456586s, CPU 0.438907s)
Info : 205441 nodes 59892 elements
Info : Writing 'le10-ref.msh'...
Info : Done writing 'le10-ref.msh'
Info : Stopped on Thu Oct 28 12:03:28 2021 (From start: Wall 1.30955s, CPU 1.44333s)
$ time feenox le10-ref.fee
sigma_y(D) = -5.3792 MPa
real 1m34.485s
user 1m30.677s
sys 0m10.449s
$This run can also be used to “calibrate” the timing. Just run the
le10-ref.fee case yourself and see how long FeenoX needs in
your server. This figure should help you to scale up (or down) the
ordinates of the figures for the parametric results shown here.
1.2 Scripted parametric execution
The driving script is called run.sh. Without any
arguments, it shows the usage:
$ ./run.sh
usage: ./run.sh { tet | hex } c_min n_steps
$ The first argument is either tet or hex.
The second is the lower end of the range for the mesh refinement
factor c \in (c_\text{min}:1]. In
principle there is no problem setting c_min to 0 because it
will never be reached exactly since the range is open to the left—
although the meshes will be insanely large. The last argument is the
number of steps.
Note that the range (c_\text{min}:1]
will be swept using a quasi-random number sequence and all the results
will be cached until removed by executing clean.sh. So if
one first runs
./run.sh tet 0.1 8and then
./run.sh tet 0.1 12the second execution would only run four actual steps, reading the
cached values for the first eight. This will hold as long as
c_min is the same for all invocations of
run.sh.
To check which of the codes are available in your configuration, run
with --check:
$ ./run.sh --check
FeenoX GAMG: yes
FeenoX MUMPS: yes
Sparselizard: yes
Code Aster: yes
CalculiX: yes
$Run a single step (i.e. c=1, which are the default meshes shown above) for each case to see if everything works.
$ ./run.sh tet 1 1
[...]
$ ./run.sh hex 1 1
[...]
$For each c both the geometry and the
mesh are created with Gmsh. The
refinements are made by setting the -clscale command-line
parameter equal to c\in
(c_\text{min}:1] to control the elements’ size. The actual values
taken by c are given by running FeenoX
with the input steps.fee. This uses a Sobol quasi-random
number sequence that starts with c=1
and then fills the interval in subsequent steps. For example, five
steps for c_\text{min} = 0.1 gives
$ feenox steps.fee 0.1 5
1
0.55
0.775
0.325
0.4375
$The first step is the coarsest mesh, which is enforced to be always
run. The second one is the middle-range mesh, and the following steps
start to “fill” in the blanks without actually reaching c=c_\text{min}. Since the run.sh
script caches the results it gets, further steps can be performed by
reusing the existing data. So if we now want to run ten steps,
$ feenox steps.fee 0.1 10
1
0.55
0.775
0.325
0.4375
0.8875
0.6625
0.2125
0.26875
0.71875
$and the first five steps will use cached data instead of re-running all the codes.
A successful execution of run.sh will give files
*.dat with the following columns:
- the parameter c \in (c_\text{min}:1]
- the total number of degrees of freedom
- the stress \sigma_y evaluated at point D
- the wall time in seconds
- the kernel-mode CPU time in seconds
- the user-mode CPU time in seconds
- the maximum memory used by the program, in kB
The rows will follow the execution order, so they will be unsorted on
the refinement factor (and number of degrees of freedom) so they are not
suitable for plotting them directly using lines to connect consecutive
data points. There is another script, report.sh that will
read the data and prepare figures in SVG format (using Gnuplot) and a markdown report with
tables with the actual figures:
$ ./report.sh tet
[...]
$ ./report.sh hex
[...]
$The report also indicates some data about the host where the test was performed and the versions of the codes used. The SVG files are interactive so they can be opened with a web browser, zoomed in and out and the individual curves can be turned on and off by clicking on the label.
2 Explanations, comments and caveats
Disclaimer: I am the author of FeenoX so all of my comments are likely to be biased. If you are reading this and feel like something is not true or is indeed way too biased, please contact me and help me to have the fairest comparison possible. There might still be some subjectivity and I apologize in advance for that.
The objective of this test is to compare consumption of resources for cloud-based computations. It is therefore suggested to run it on a cloud server and not on a local laptop.
In order to have the most fair comparison possible, even though the codes can measure CPU and memory consumption themselves, all of them are run through the
timetool (the actual binary tool at/usr/bin/time, not the shell’s internal).This is a serial test only so the variable
OMP_NUM_THREADSis set to one to avoid spawning OpenMP threads. MPI-based parallel tests across different hosts will come later on. OpenMP will not be the main focus of the scalability study.The wall time should thus be equal to the sum of kernel-space CPU time plus user-space CPU time plus some latency that depends on the operating system’s scheduler.
The hex mesh is created as a first-order mesh and then either
- converted to a (straight) second-order incomplete (i.e. hex20) mesh, or
- the first-order mesh is fed to the code and it is asked to use second-order elements.
depending on the capabilities of each code (some codes honor the element type in the mesh but others change the order and number of degrees of freedom per element on the fly).
The second column of the output is the total number of degrees of freedom. In principle for a simple three-dimensional problem like this one it should be equal to three times the number of nodes. But by default Code Aster sets Dirichlet boundary conditions as Lagrange multipliers, increasing the matrix size. On the contrary, CalculiX removes the degrees of freedom that correspond to nodes with Dirichlet boundary conditions resulting in a smaller matrix size. Sparselizard needs complete elements so for order = 2 it assigns an unknown to each node, edge, face and volume of the element, resulting in 27 unknowns per hexahedron. This is equivalent of using hex27 (instead of hex20) elements, resulting in a (much) higher degree-of-freedom count for the hexahedral case.
Most codes allow to choose the actual linear sparse solver at runtime, maybe depending on the availability of particular solving libraries at compilation time. For these codes, there are many curves, one for each supported preconditioner+solver combination.
When the mesh is big, chances are that the code runs out of memory being killed by the operating system (if there is no swap partition, which there should be not for efficiency reasons). Code Aster and CalculiX have out-of-core capabilities in which they can run with less memory than the actual requested, at the expense of larger CPU times by using a tailor-made disk-swapping procedure. The other codes are killed by the operating system when this happens and there is no data point for that particular value of c.
The number of iterations (and thus the CPU time) needed to converge when using iterative solvers depends on the tolerance. In all cases, default tolerances have been used.
2.1 FeenoX
The run.sh script calls the executable
feenox with the le10.fee input file as the
first argument and ${m}-${c} as the second one, resulting
in something like tet-1 or hex-0.2. This
argument is expanded where the input file contains a $1,
namely the mesh file name. FeenoX prints the total number of degrees of
freedom and the stress \sigma_y at
point D. It does not write any
post-processing file (the WRITE_MESH keyword is commented
out):
# NAFEMS Benchmark LE-10: thick plate pressure
PROBLEM mechanical
READ_MESH le10_2nd-$1.msh # FeenoX honors the order of the mesh
BC upper p=1 # 1 Mpa
BC DCDC v=0 # Face DCD'C' zero y-displacement
BC ABAB u=0 # Face ABA'B' zero x-displacement
BC BCBC u=0 v=0 # Face BCB'C' x and y displ. fixed
BC midplane w=0 # z displacements fixed along mid-plane
E = 210e3 # Young modulus in MPa (because mesh is in mm)
nu = 0.3 # Poisson's ratio
SOLVE_PROBLEM # TODO: implicit
PRINT total_dofs %.8f sigmay(2000,0,300)
# write post-processing data for paraview
# WRITE_MESH le10-feenox-${c}.vtk VECTOR u v w sigmax sigmay sigmaz tauxy tauzx tauyzThe mesh file should already contain a second-order mesh. The file
le10-tet.geo creates tet10 elements, but the
le10-hex.geo creates hex8 elements that have to be
converted to hex20 before FeenoX can use them. This is achieved in the
run.sh script within the block
if [ ! -e le10-${m}-${c}.msh ]; then
gmsh -3 le10-${m}.geo -clscale ${c} -o le10-${m}-${c}.msh || exit 1
gmsh -3 le10-${m}-${c}.msh -setnumber Mesh.SecondOrderIncomplete 1 -order 2 -o le10_2nd-${m}-${c}.msh || exit 1
fiIf the file le10-${m}-${c}.msh does not exist (otherwise
it would take it as a cached file), Gmsh is first called with either
le10-tet.geo or le10-hex.geo as the input and
-clscale is set to c to
create le10-${m}-${c}.msh. Then, this mesh file is
explicitly converted to a second-order mesh with -order 2
and any hex8 is converted to hex20 (instead of hex27 because
Mesh.SecondOrderIncomplete=1) and saved as
le10_2nd-${m}-${c}.msh. It is this last mesh file the one
that FeenoX needs.
By default, FeenoX uses Mark Adams’ Geometric-Algebraic
Multigrid Preconditioner provided by PETSc and conjugate
gradients as the iterative solver for the mechanical problem. This
default corresponds to the “feenox_gamg” curve. One can check the actual
options by passing --ksp_view as an extra option, e.g.
$ feenox le10.fee tet-1 --ksp_view
KSP Object: 1 MPI processes
type: cg
maximum iterations=10000, nonzero initial guess
tolerances: relative=1e-06, absolute=1e-50, divergence=10000.
left preconditioning
using PRECONDITIONED norm type for convergence test
PC Object: 1 MPI processes
type: gamg
type is MULTIPLICATIVE, levels=4 cycles=v
Cycles per PCApply=1
Using externally compute Galerkin coarse grid matrices
[...]
linear system matrix = precond matrix:
Mat Object: K_bc 1 MPI processes
type: seqaij
rows=20325, cols=20325, bs=3
total: nonzeros=1498671, allocated nonzeros=1498671
total number of mallocs used during MatSetValues calls=0
has attached near null space
using I-node routines: found 6775 nodes, limit used is 5
20325 -5.41620113
$If the MUMPS
solver is available through PETSc, run.sh adds the
command-line option --mumps to create another curve,
“feenox_mumps”:
$ feenox le10.fee tet-1 --mumps --ksp_view
KSP Object: 1 MPI processes
type: preonly
maximum iterations=10000, initial guess is zero
tolerances: relative=1e-06, absolute=1e-50, divergence=10000.
left preconditioning
using NONE norm type for convergence test
PC Object: 1 MPI processes
type: cholesky
out-of-place factorization
tolerance for zero pivot 2.22045e-14
matrix ordering: nd
factor fill ratio given 0., needed 0.
Factored matrix follows:
Mat Object: 1 MPI processes
type: mumps
rows=20325, cols=20325
package used to perform factorization: mumps
total: nonzeros=5974629, allocated nonzeros=5974629
MUMPS run parameters:
[...]
INFOG(39) (after analysis: estimated size of all MUMPS internal data for running BLR out-of-core - sum over all processors): 0
linear system matrix = precond matrix:
Mat Object: K_bc 1 MPI processes
type: seqaij
rows=20325, cols=20325, bs=3
total: nonzeros=1498671, allocated nonzeros=1498671
total number of mallocs used during MatSetValues calls=0
using I-node routines: found 6775 nodes, limit used is 5
20325 -5.41600874
$ 2.2 Sparselizard
Thanks to Alexandre Halbach for the discussions about Sparselizard’s
internals. The following main.cpp is used to solve the
NAFEMS LE10 benchmark with Sparselizard:
// NAFEMS LE10 Benchmark solved with Sparselizard
#include "sparselizard.h"
using namespace sl;
int main(int argc, char **argv) {
int bulk = 1;
int upper = 2;
int DCDC = 3;
int ABAB = 4;
int BCBC = 5;
int midplane = 6;
double young = 210e3;
double poisson = 0.3;
std::string c = (argc > 1) ? argv[1] : "tet-1";
mesh mymesh("gmsh:../le10-"+c+".msh", 0);
field u("h1xyz");
parameter E;
E|bulk = young;
parameter nu;
nu|bulk = poisson;
u.setorder(bulk, 2);
u.compy().setconstraint(DCDC); // v=0 @ DCDC
u.compx().setconstraint(ABAB); // u=0 @ ABAB
u.compx().setconstraint(BCBC); // u=0 @ BCBC
u.compy().setconstraint(BCBC); // v=0 @ BCBC
u.compz().setconstraint(midplane); // w=0 @ midplae
formulation elasticity;
elasticity += integral(upper, array1x3(0,0,-1)*tf(u)); // p=1 @ upper
elasticity += integral(bulk, predefinedelasticity(dof(u), tf(u), E, nu), -2); // -2 gives an exact integration for up to 4th order polynomial"
elasticity.generate();
vec solu = solve(elasticity.A(), elasticity.b(), "cholesky");
// Transfer the data from the solution vector to the u field:
u.setdata(bulk, solu);
double lambda = young * poisson/((1+poisson)*(1-2*poisson));
double mu = 0.5*young/(1+poisson);
expression H(6,6,{lambda+2*mu, lambda, lambda, 0, 0, 0,
lambda, lambda+2*mu, lambda, 0, 0, 0,
lambda, lambda, lambda+2*mu, 0, 0, 0,
0, 0, 0, mu, 0, 0,
0, 0, 0, 0, mu, 0,
0, 0, 0, 0, 0, mu});
expression sigma = H*strain(u);
// u.write(bulk, "le10-sparselizard-displ.vtk", 2);
// comp(1, sigma).write(bulk, "le10-sparselizard-sigmay.vtk", 2);
field sigmayy("h1");
sigmayy.setorder(bulk, 2);
sigmayy.setvalue(bulk, comp(1, sigma));
std::cout << elasticity.countdofs() << "\t" << sigmayy.interpolate(bulk, {2000, 0, 300})[0] << std::endl;
return 0;
}
The main function takes one argument which should be the same as in
FeenoX, i.e. tet-1, hex-0.25, etc. which is
used to read the mesh file as created by Gmsh from the parent directory.
The problem order is set to two. The Dirichlet BCs are then set. The
Neumann BC is set into the elasticity weak formulation as a surface
integral. The volume integral is performed using 2nd-order Gauss points
and then the problem is solved for the displacements. The stress tensor
field is explicitly computed out of the strain using the linear elastic
6x6 matrix in Voigt notation. The stress at point D is interpolated from a smoothed field over
all the elemental contributions to the node and printed into the
standard output, along with the total number of degrees of freedom being
solved for. Sparselizard uses MUMPS through PETSc. By default, it will
use the LU preconditioner. But since the stiffness matrix is symmetric,
we choose to use the Cholesky preconditioner.
Note that as already discussed, Sparselizards needs tensor-product elements for the hex case. Therefore, the choice of order equal to two triggers the addition of unknowns at the 12 edges, at the 6 faces and 1 at the volume besides the 8 corners resulting in 27 unknowns (per each of the three degrees of freedom of the problem). The other codes stick to incomplete hex20 elements, so the total number of degrees of freedom is larger for Sparselizard than for the other codes for the same c.
2.3 Code Aster
Thanks to Cyprien Rusu for all the help setting up the code and the input files and to Nicolas Tardieu for an interesting technical discussion about Code Aster’s internals.
Even though Gmsh can write the mesh in the very efficient MED format which Code Aster can read, since it is a binary file (it is based on HDF5) the version of the MED library which both Gmsh as Aster are linked with should be the same. Since Code Aster is tricky to compile with custom dependencies and Gmsh uses a newer MED library by default, this option was dismissed.
It was decided to use the text-based (and archaic format UNV). The
second-order mesh le10_2nd-${m}-${c}.msh is converted to
UNV with Gmsh:
if [ ! -e le10_2nd-${m}-${c}.unv ]; then
gmsh -3 le10_2nd-${m}-${c}.msh -o le10_2nd-${m}-${c}.unv || exit 1
fiThe argument that the Code Aster executable needs in order to run a
case is an “export” file that defines some run-time options for the
execution (memory and CPU limits, number of MPI instances, etc.) and
links Fortran file units (the ones that were introduced in 1954) to
actual file system names, like the “comm” (input) file, the mesh file,
the output file, etc. Interestingly enough, Code Aster would modify the
input export file (sic) and rename relative file paths contained in it
to absolute ones. This makes it hard to track export files with Git, but
what run.sh does is it uses a template export file
P actions make_etude
P debug nodebug
P memjob 2097152
P memory_limit 15500.0
P mode interactif
P mpi_nbcpu 1
P mpi_nbnoeud 1
P ncpus 0
P time_limit 9000.0
P tpsjob 16
P version stable
A memjeveux 128.0
A tpmax 9000.0
F comm le10__s_.comm D 1
F libr le10_2nd-_m_.unv D 20
F libr le10-_m_.rmed R 80
F mess message__s_-_m_ R 6
F resu DD-_s_-_m_.txt R 17
R base base-stage1-_s_-_m_ R 0
and then use sed would replace _m_ with
${m} to have a per-c
Git-ignored export file which can be further modified as needed.
The actual problem definition is stored in a “comm” file. There are three variants to solve the NAFEMS LE10 problem with Code Aster, namely
le10_default.commDEBUT(LANG='EN') mesh = LIRE_MAILLAGE(UNITE=20, FORMAT='IDEAS') model = AFFE_MODELE(AFFE=_F(MODELISATION=('3D', ), PHENOMENE='MECANIQUE', TOUT='OUI'), MAILLAGE=mesh) mater = DEFI_MATERIAU(ELAS=_F(E=210000.0, NU=0.3)) fieldmat = AFFE_MATERIAU(AFFE=_F(MATER=(mater, ), TOUT='OUI'), MODELE=model) load = AFFE_CHAR_MECA(DDL_IMPO=(_F(DY=0.0, GROUP_MA=('DCDC', )), _F(DX=0.0, GROUP_MA=('ABAB', )), _F(DX=0.0, DY=0.0, GROUP_MA=('BCBC', )), _F(DZ=0.0, GROUP_MA=('midplane', ))), MODELE=model, PRES_REP=_F(GROUP_MA=('upper', ), PRES=1.0)) reslin = MECA_STATIQUE(CHAM_MATER=fieldmat, EXCIT=_F(CHARGE=load), MODELE=model) reslin = CALC_CHAMP(reuse=reslin, CONTRAINTE=('SIGM_ELGA', 'SIGM_ELNO', 'SIGM_NOEU', 'SIEF_ELGA', 'SIEF_ELNO', 'SIEF_NOEU'), CRITERES=('SIEQ_ELGA', 'SIEQ_ELNO', 'SIEQ_NOEU'), RESULTAT=reslin) #IMPR_RESU(RESU=_F(RESULTAT=reslin), UNITE=80) IMPR_RESU( FORMAT='RESULTAT', RESU=_F( GROUP_MA=('DD', ), IMPR_COOR='OUI', NOM_CHAM=('SIGM_NOEU', ), NOM_CMP=('SIYY'), RESULTAT=reslin ), UNITE=17 ) FIN()le10_cholesky.commDEBUT(LANG='EN') mesh = LIRE_MAILLAGE(UNITE=20, FORMAT='IDEAS') model = AFFE_MODELE(AFFE=_F(MODELISATION=('3D', ), PHENOMENE='MECANIQUE', TOUT='OUI'), MAILLAGE=mesh) mater = DEFI_MATERIAU(ELAS=_F(E=210000.0, NU=0.3)) fieldmat = AFFE_MATERIAU(AFFE=_F(MATER=(mater, ), TOUT='OUI'), MODELE=model) BC = AFFE_CHAR_CINE(MECA_IMPO=(_F(DY=0.0, GROUP_MA=('DCDC', )), _F(DX=0.0, GROUP_MA=('ABAB', )), _F(DX=0.0, DY=0.0, GROUP_MA=('BCBC', )), _F(DZ=0.0, GROUP_MA=('midplane', ))), MODELE=model, ) load = AFFE_CHAR_MECA( MODELE=model, PRES_REP=_F(GROUP_MA=('upper', ), PRES=1.0)) reslin = MECA_STATIQUE(CHAM_MATER=fieldmat, EXCIT=(_F(CHARGE=BC),_F(CHARGE=load),), MODELE=model, SOLVEUR=_F(METHODE='GCPC', PRE_COND='LDLT_INC'), ) reslin = CALC_CHAMP(reuse=reslin, CONTRAINTE=('SIGM_ELGA', 'SIGM_ELNO', 'SIGM_NOEU', 'SIEF_ELGA', 'SIEF_ELNO', 'SIEF_NOEU'), CRITERES=('SIEQ_ELGA', 'SIEQ_ELNO', 'SIEQ_NOEU'), RESULTAT=reslin) #IMPR_RESU(RESU=_F(RESULTAT=reslin), UNITE=80) IMPR_RESU( FORMAT='RESULTAT', RESU=_F( GROUP_MA=('DD', ), IMPR_COOR='OUI', NOM_CHAM=('SIGM_NOEU', ), NOM_CMP=('SIYY'), RESULTAT=reslin ), UNITE=17 ) FIN()le10_mumps.commDEBUT(LANG='EN') mesh = LIRE_MAILLAGE(UNITE=20, FORMAT='IDEAS') model = AFFE_MODELE(AFFE=_F(MODELISATION=('3D', ), PHENOMENE='MECANIQUE', TOUT='OUI'), MAILLAGE=mesh) mater = DEFI_MATERIAU(ELAS=_F(E=210000.0, NU=0.3)) fieldmat = AFFE_MATERIAU(AFFE=_F(MATER=(mater, ), TOUT='OUI'), MODELE=model) BC = AFFE_CHAR_CINE(MECA_IMPO=(_F(DY=0.0, GROUP_MA=('DCDC', )), _F(DX=0.0, GROUP_MA=('ABAB', )), _F(DX=0.0, DY=0.0, GROUP_MA=('BCBC', )), _F(DZ=0.0, GROUP_MA=('midplane', ))), MODELE=model, ) load = AFFE_CHAR_MECA( MODELE=model, PRES_REP=_F(GROUP_MA=('upper', ), PRES=1.0)) reslin = MECA_STATIQUE(CHAM_MATER=fieldmat, EXCIT=(_F(CHARGE=BC),_F(CHARGE=load),), MODELE=model, SOLVEUR=_F(METHODE='MUMPS',)) reslin = CALC_CHAMP(reuse=reslin, CONTRAINTE=('SIGM_ELGA', 'SIGM_ELNO', 'SIGM_NOEU', 'SIEF_ELGA', 'SIEF_ELNO', 'SIEF_NOEU'), CRITERES=('SIEQ_ELGA', 'SIEQ_ELNO', 'SIEQ_NOEU'), RESULTAT=reslin) #IMPR_RESU(RESU=_F(RESULTAT=reslin), UNITE=80) IMPR_RESU( FORMAT='RESULTAT', RESU=_F( GROUP_MA=('DD', ), IMPR_COOR='OUI', NOM_CHAM=('SIGM_NOEU', ), NOM_CMP=('SIYY'), RESULTAT=reslin ), UNITE=17 ) FIN()
To retrieve the stress at point D,
instead of writing the full results into unit 80
(i.e. le10-${m}.rmed), only an ASCII file with nodal values
of the stress tensor are written for those nodes that belong to the
DD physical group (the D-D^\prime)
segment in the original geometry. For some reason, Code Aster won’t
accept a zero-dimensional physical group (i.e. a point) to write the
ASCII result. So the run.sh has to parse this ASCII file so
as to find the row corresponding to point D and extract the value of \sigma_y, which is the fifth column:
grep "2.00000000000000E+03 0.00000000000000E+00 3.00000000000000E+02" DD-${m}-${c}.txt | awk '{print $5}' >> aster_default_${m}-${c}.sigmayThe total number of degrees of freedom is taken from the “message” output (unit 6) by grepping the French expression for “degrees of freedom”. Luckily UTF-8 works well:
grep "degrés de liberté:" message-${m}-${c} | awk '{printf("%g\t", $7)}' > aster_default_${m}-${c}.sigmay2.4 CalculiX
Thanks Sergio Pluchinsky for all the help to set up the inputs and the mesh files.
As explained below, I just followed his suggestions without understanding them. The way CalculiX input files work will remain unheard of to me for the time being.
The following paragraph explains what run.sh does (which
is needlessly cumbersome IMHO) to make CalculiX work. It should be noted
before the explanation starts that I had to modify the Gmsh UNV writer
to handle both “groups of nodes” and “groups of elements” at the same
time: https://gitlab.onelab.info/gmsh/gmsh/-/commit/a7fef9f6e8a7c870cf39b8702c57f3e33bfa948d.
So make sure the Gmsh version used is later than that commit. Also,
there is this unical.c conversion tool from UNV to INP that
I initially borrowed from https://github.com/calculix/unical1 but had to modify to
make it work. The code is included in the test directory and compiled
when run.sh detects CalculiX is available. For large
problems, this conversion procedure takes a non-trivial amount of time
(i.e. more than five minutes) which is not accounted for in the
resulting curves.
The second-order mesh le10_2nd-${m}-${c}.msh is
converted to UNV with both options SaveGroupsOfElements and
SaveGroupsOfNodes equal to true. Then, this UNV (which as
already explained, needs a Gmsh version later than commit
a7fef9f6 from November 2021 otherwise the next step will
fail if there are an odd number of nodes) is read by the
slightly-modified tool unica1l that creates a
.inp mesh file which can be read by CalculiX:
if [ ! -e le10_mesh-${m}-${c}.inp ]; then
gmsh -3 le10_2nd-${m}-${c}.msh -setnumber Mesh.SaveGroupsOfElements 1 -setnumber Mesh.SaveGroupsOfNodes 1 -o le10_calculix-${m}-${c}.unv || exit 1
./unical1 le10_calculix-${m}-${c}.unv le10_mesh-${m}-${c}.inp || exit
fiThe main input is a template that reads the appropriate
.inp mesh file for c and
sets successively the Spooles solver (“calculix_spooles”), the internal
with diagonal scaling (“calculix_diagonal”) and the internal with
Cholesky preconditioning (“calculix_cholesky”). There is a reason I
still cannot understand that explains why there has to be one template
for tet and one for hex. In effect, Sergio
pointed me to section that explains the DLOAD keyword (used
to set a pressure boundary condition) in the ccx manual. It
says (casing is verbatim):
This option allows the specification of distributed loads. These include constant pressure loading on element faces, edge loading on shells and mass loading (load per unit mass) either by gravity forces or by centrifugal forces. For surface loading the faces of the elements are numbered as follows (for the node numbering of the elements see Section 3.1):
for hexahedral elements:
- face 1: 1-2-3-4
- face 2: 5-8-7-6
- face 3: 1-5-6-2
- face 4: 2-6-7-3
- face 5: 3-7-8-4
- face 6: 4-8-5-1
for tetrahedral elements:
- Face 1: 1-2-3
- Face 2: 1-4-2
- Face 3: 2-4-3
- Face 4: 3-4-1
So it seems that CalculiX needs some sort of mesh-dependent numbering
of the faces of volumetric elements to set surface Neumann boundary
conditions (!). This is why I had to blindly rely on Sergio’s expertise
to handle these UPPERFn and Pn lines within
the Dload section below. In any case, the two templates for
tets and hexes are, respectively:
le10-tet.inp*include, input = le10_mesh-tet-xxx.inp *Material, Name=STEEL *Elastic 200e3, 0.3 *Solid section, Elset=C3D10, Material=STEEL *Step **Static, Solver=PaStiX **Static, Solver=Pardiso **Static, Solver=Spooles **Static, Solver=Iterative scaling **Static, Solver=Iterative Cholesky *Boundary, Fixed ABAB, 1, 1 DCDC, 2, 2 BCBC, 1, 2 MIDPLANE, 3, 3 *Dload UPPERF1, P1, 1 UPPERF2, P2, 1 UPPERF3, P3, 1 UPPERF4, P4, 1 *Node file RF, U *El file S, E *End steple10-hex.inp*include, input = le10_mesh-hex-xxx.inp *Material, Name=STEEL *Elastic 200e3, 0.3 *Solid section, Elset=C3D20, Material=STEEL *Step **Static, Solver=PaStiX **Static, Solver=Pardiso **Static, Solver=Spooles **Static, Solver=Iterative scaling **Static, Solver=Iterative Cholesky *Boundary, Fixed ABAB, 1, 1 DCDC, 2, 2 BCBC, 1, 2 MIDPLANE, 3, 3 *Dload UPPERF6, P6, 1 *Node file RF, U *El file S, E *End step
These templates are filtered with sed that replaces
xxx with the appropriate mesh and un-comments each of the
solver lines successively to have a working input file:
sed s/xxx/${c}/ le10-${m}.inp | sed 's/**Static, Solver=Spooles/*Static, Solver=Spooles/' > le10_spooles_${m}-${c}.inpTo read the stress at point D, an
awk file that parses the output .frd file and
searches for the nodal values of the stress tensor based on the
coordinates of the point D had to be
written. It has to take into account that this .frd file
does not have blank-separated fields but fixed-width ASCII columns, such
as
-1 371-1.52894E+00 1.19627E+00-5.85388E-02-8.97020E-03 1.73081E-03 7.02901E-01where negative values appear concatenated with the previous one as a single ASCII token in a non-UNIX-friendly way.
#!/usr/bin/gawk
{
# this only works for all-positive coordinates, otherwise we would have to use substr()
if ($3 == "2.00000E+03" && $4 = "0.00000E+00" && $5 == "3.00000E+02")
{
node = $2
}
if ($1 == -4 && $2 == "STRESS") {
stresses = 1
}
if (node != 0 && stresses == 1 && found == 0) {
if (strtonum(substr($0,4,10)) == node) {
printf("%e\t", substr($0, 26, 12))
found = 1
}
}
}The total number of degrees of freedom is read from the standard output grepping for “number of equations”:
grep -C 1 "number of equations" calculix_spooles_${m}-${c}.txt | tail -n 1 | awk '{printf("%d\t", $1)}' > calculix_spooles_${m}-${c}.sigmay2.5 Reflex
Reflex is OnScale’s next-generation finite element software framework architected for maximum performance, scalability, and programming efficiency.
3 Setting up the codes
3.1 Gmsh
Both the continuous geometry and the discretized meshes are created
with Gmsh. The run.sh
script will not run if gmsh is not a valid command.
It is better to use latest versions instead of the one distributed in the operating system’s package repositories. In fact, to run the CalculiX test, version 4.9 or later is needed. Either the official binaries or a compiled-from-scratch version will do.
The no-X binary version can be used. To download and copy it to a system-wide location do:
wget http://gmsh.info/bin/Linux/gmsh-nox-git-Linux64-sdk.tgz
tar xvzf gmsh-nox-git-Linux64-sdk.tgz
sudo cp gmsh-nox-git-Linux64-sdk/bin/gmsh /usr/local/bin
sudo cp -P gmsh-nox-git-Linux64-sdk/lib/* /usr/local/libAlternatively, it can be compile from source (OpenCASCADE is needed to create the LE10’s CAD):
sudo apt-get install libocct-data-exchange-dev libocct-foundation-dev libocct-modeling-data-dev
git clone https://gitlab.onelab.info/gmsh/gmsh.git
cd gmsh
mkdir build && cd build
cmake ..
make
sudo make installCheck it works globally:
$ gmsh -info
Version : 4.9.0-nox-git-701db57af
License : GNU General Public License
Build OS : Linux64-sdk
Build date : 20211019
Build host : gmsh.info
Build options : 64Bit ALGLIB ANN Bamg Blas[petsc] Blossom Cgns DIntegration Dlopen DomHex Eigen Gmm Hxt Kbipack Lapack[petsc] LinuxJoystick MathEx Med Mesh Metis Mmg Netgen ONELAB ONELABMetamodel OpenCASCADE OpenMP OptHom PETSc Parser Plugins Post QuadMeshingTools QuadTri Solver TetGen/BR Voro++ WinslowUntangler Zlib
PETSc version : 3.14.4 (real arithmtic)
OCC version : 7.6.0
MED version : 4.1.0
Packaged by : geuzaine
Web site : https://gmsh.info
Issue tracker : https://gitlab.onelab.info/gmsh/gmsh/issues
$3.2 FeenoX
Not only is FeenoX the
docus of the current test, but it also is used to compute the
quasi-random sequence of mesh refinements factors c. So it is mandatory to have a working
feenox command.
The easiest way to set up FeenoX is to download, un-compress and copy a statically-linked binary to a system-wide location:
wget https://seamplex.com/feenox/dist/linux/feenox-v0.1.152-g8329396-linux-amd64.tar.gz
tar xvzf feenox-v0.1.152-g8329396-linux-amd64.tar.gz
sudo cp feenox-v0.1.152-g8329396-linux-amd64/bin/feenox /usr/local/binInstead, it can be compiled from the Github repository using
stack PETSc from apt (which might be considered “old” in
some GNU/Linux distributions). This will also enable the MUMPS solver
(so an the extra curve named “feenox_mumps” will be added to the
results):
sudo apt-get install gcc make git automake autoconf libgsl-dev petsc-dev slepc-dev
git clone https://github.com/seamplex/feenox
cd feenox
./autogen.sh
./configure
make
make check
sudo make installCustom PETSc versions and architectures are supported as well. The
default preconditioner+solver pair gamg+cg is supported with all PETSc
configurations. To make the MUMPS direct solver available, PETSc has to
be configured and linked properly (i.e. configure with
--download-mumps or use the PETSc packages from the
operating system’s repositories). See the compilation
guide for further details.
Either way, check it works globally:
$ feenox -V
FeenoX v0.1.159-gab7abd8-dirty
a free no-fee no-X uniX-like finite-element(ish) computational engineering tool
Last commit date : Thu Oct 28 10:43:38 2021 -0300
Build date : Thu Oct 28 13:52:11 2021 +0000
Build architecture : linux-gnu x86_64
Compiler : gcc (Ubuntu 9.3.0-17ubuntu1~20.04) 9.3.0
Compiler flags : -O3
Builder : ubuntu@ip-172-31-44-208
GSL version : 2.5
SUNDIALS version : 3.1.2
PETSc version : Petsc Release Version 3.12.4, Feb, 04, 2020
PETSc arch :
PETSc options : --build=x86_64-linux-gnu --prefix=/usr --includedir=${prefix}/include --mandir=${prefix}/share/man --infodir=${prefix}/share/info --sysconfdir=/etc --localstatedir=/var --with-silent-rules=0 --libdir=${prefix}/lib/x86_64-linux-gnu --runstatedir=/run --with-maintainer-mode=0 --with-dependency-tracking=0 --with-debugging=0 --shared-library-extension=_real --with-shared-libraries --with-pic=1 --with-cc=mpicc --with-cxx=mpicxx --with-fc=mpif90 --with-cxx-dialect=C++11 --with-opencl=1 --with-blas-lib=-lblas --with-lapack-lib=-llapack --with-scalapack=1 --with-scalapack-lib=-lscalapack-openmpi --with-mumps=1 --with-mumps-include="[]" --with-mumps-lib="-ldmumps -lzmumps -lsmumps -lcmumps -lmumps_common -lpord" --with-suitesparse=1 --with-suitesparse-include=/usr/include/suitesparse --with-suitesparse-lib="-lumfpack -lamd -lcholmod -lklu" --with-ptscotch=1 --with-ptscotch-include=/usr/include/scotch --with-ptscotch-lib="-lptesmumps -lptscotch -lptscotcherr" --with-fftw=1 --with-fftw-include="[]" --with-fftw-lib="-lfftw3 -lfftw3_mpi" --with-superlu=1 --with-superlu-include=/usr/include/superlu --with-superlu-lib=-lsuperlu --with-superlu_dist=1 --with-superlu_dist-include=/usr/include/superlu-dist --with-superlu_dist-lib=-lsuperlu_dist --with-hdf5-include=/usr/include/hdf5/openmpi --with-hdf5-lib="-L/usr/lib/x86_64-linux-gnu/hdf5/openmpi -L/usr/lib/openmpi/lib -lhdf5 -lmpi" --CXX_LINKER_FLAGS=-Wl,--no-as-needed --with-hypre=1 --with-hypre-include=/usr/include/hypre --with-hypre-lib=-lHYPRE_core --prefix=/usr/lib/petscdir/petsc3.12/x86_64-linux-gnu-real --PETSC_ARCH=x86_64-linux-gnu-real CFLAGS="-g -O2 -fstack-protector-strong -Wformat -Werror=format-security -fPIC" CXXFLAGS="-g -O2 -fstack-protector-strong -Wformat -Werror=format-security -fPIC" FCFLAGS="-g -O2 -fstack-protector-strong -fPIC -ffree-line-length-0" FFLAGS="-g -O2 -fstack-protector-strong -fPIC -ffree-line-length-0" CPPFLAGS="-Wdate-time -D_FORTIFY_SOURCE=2" LDFLAGS="-Wl,-Bsymbolic-functions -Wl,-z,relro -fPIC" MAKEFLAGS=w
SLEPc version : SLEPc Release Version 3.12.2, Jan 13, 2020
$The PETSc options line will tell if MUMPS is available
or not, so grepping will tell:
$ feenox -V | grep -i mumps | wc -l
1
$3.3 Sparselizard
In order to test Sparselizard,
a sub-directory named sparselizard should exist in the
directory where run.sh is, and an executable named
sparselizard should exist in that sub-directory.
This can be achieved by cloning and compiling the Github repository. It
needs a particularly-configured PETSc/SLEPc version, which can be
obtained by executing the
install_external_libs/install_petsc.sh script (which pulls
latest PETSc main from the Gitlab repository, configures and compiles
it). Also, the Gmsh API is needed to read .msh v4, so run
install_external_libsoptional_install_gmsh_api.sh as well.
The main.cpp is the file le10.cpp provided in
the le10-aster-lizard directory. Therefore, a symbolic link
has to be added in the sparselizard directory to point
main.cpp to ../le10.cpp:
cd feenox/tests/nafems/le10
git clone https://github.com/halbux/sparselizard/
cd sparselizard/
ln -s ../le10.cpp main.cpp
cd install_external_libs/
./install_petsc.sh
./optional_install_gmsh_api.sh
cd ..
make
cd ..3.4 Code Aster
Code Aster is tricky to compile (and use, at least for me). The following works (only) in Ubuntu 20.04:
wget https://www.code-aster.org/FICHIERS/aster-full-src-14.6.0-1.noarch.tar.gz
tar xvzf aster-full-src-14.6.0-1.noarch.tar.gz
cd aster-full-src-14.6.0
mkdir -p $HOME/aster
python3 setup.py --aster_root=$HOME/aster
cd
source aster/etc/codeaster/profile.sh Check it does work globally:
$ as_run --getversion
<INFO> Version exploitation 14.6.0 - 11/06/2020 - rev. b3490fa3b76c
$Note that compilation from the repository fails in Debian 11 with gcc10:
$ sudo apt-get install mercurial
$ sudo apt-get install gcc g++ gfortran cmake python3 python3-dev python3-numpy tk bison flex dh-exec
$ sudo apt-get install liblapack-dev libblas-dev libboost-python-dev libboost-numpy-dev zlib1g-dev
$ hg clone http://hg.code.sf.net/p/codeaster/src codeaster-src
$ cd codeaster-src
$ ./waf configure
$ ./waf build
checking environment... executing: ./waf.engine build --out=build/std --jobs=4
Waf: Entering directory `/home/gtheler/codigos/3ros/codeaster-src/build/std/release'
[1521/8549] Processing bibfor/echange/lub_module.F90
[2174/8549] Compiling bibfor/prepost/cmqlql.F90
[2175/8549] Compiling bibfor/prepost/cmhho.F90
[2177/8549] Compiling bibfor/prepost/cm1518.F90
[4370/8549] Compiling bibfor/algorith/dtmprep_noli_lub.F90
[6623/8549] Compiling bibfor/algorith/dtmclean_noli_yacs.F90
[7258/8549] Compiling bibcxx/Utilities/GenericParameter.cxx
[7259/8549] Compiling bibcxx/Utilities/CppToFortranGlossary.cxx
[7260/8549] Compiling bibcxx/Utilities/ConvertibleValue.cxx
[7263/8549] Compiling bibcxx/PythonBindings/LoadResultInterface.cxx
In file included from ../../../bibcxx/Utilities/ConvertibleValue.cxx:24:
../../../bibcxx/Utilities/ConvertibleValue.h: In member function ‘const ReturnValue& ConvertibleValue<ValueType1, ValueType2>::getValue() const’:
../../../bibcxx/Utilities/ConvertibleValue.h:69:24: error: ‘runtime_error’ is not a member of ‘std’
69 | throw std::runtime_error( "Impossible to convert " + _valToConvert );
| ^~~~~~~~~~~~~
In file included from /usr/include/boost/smart_ptr/detail/sp_thread_sleep.hpp:22,
from /usr/include/boost/smart_ptr/detail/yield_k.hpp:23,
from /usr/include/boost/smart_ptr/detail/spinlock_gcc_atomic.hpp:14,
from /usr/include/boost/smart_ptr/detail/spinlock.hpp:42,
from /usr/include/boost/smart_ptr/detail/spinlock_pool.hpp:25,
from /usr/include/boost/smart_ptr/shared_ptr.hpp:29,
from /usr/include/boost/shared_ptr.hpp:17,
from ../../../bibcxx/include/astercxx.h:37,
from ../../../bibcxx/PythonBindings/LoadResultInterface.h:27,
from ../../../bibcxx/PythonBindings/LoadResultInterface.cxx:24:
/usr/include/boost/bind.hpp:36:1: note: ‘#pragma message: The practice of declaring the Bind placeholders (_1, _2, ...) in the global namespace is deprecated. Please use <boost/bind/bind.hpp> + using namespace boost::placeholders, or define BOOST_BIND_GLOBAL_PLACEHOLDERS to retain the current behavior.’
36 | BOOST_PRAGMA_MESSAGE(
| ^~~~~~~~~~~~~~~~~~~~
/usr/include/boost/detail/iterator.hpp:13:1: note: ‘#pragma message: This header is deprecated. Use <iterator> instead.’
13 | BOOST_HEADER_DEPRECATED("<iterator>")
| ^~~~~~~~~~~~~~~~~~~~~~~
Waf: Leaving directory `/home/gtheler/codigos/3ros/codeaster-src/build/std/release'
Build failed
-> task in 'asterbibcxx' failed with exit status 1 (run with -v to display more information)
$ 3.5 CalculiX
Thanks Sergio Pluchinsky for all the help to set up the inputs and the mesh files.
CalculiX is available at Debian/Ubuntu repositories, although the versions are not up to date and they only have the Spooles solver and the internal iterative solver with either diagonal or Cholesky preconditioning.
There are sources which come with an already-working makefile (i.e. the don’t need configuration). Sadly, the official sources won’t compile (and throw millions of warnings) in Debian 11 with gcc10:
$ wget http://www.dhondt.de/ccx_2.18.src.tar.bz2
$ tar xf ccx_2.18.src.tar.bz2
$ cd CalculiX/ccx_2.18/src/
$ make
[...]
21 | & nodef,idirf,df,cp,r,physcon,numf,set,mi,ider,ttime,time,
| 1
Warning: Unused dummy argument ‘ttime’ at (1) [-Wunused-dummy-argument]
cross_split.f:101:72:
101 | & *(1.d0-pt2pt1**(1.d0/kdkm1))/r)/dsqrt(Tt1)
| ^
Warning: ‘a’ may be used uninitialized in this function [-Wmaybe-uninitialized]
gfortran -Wall -O2 -c cubic.f
gfortran -Wall -O2 -c cubtri.f
cubtri.f:131:18:
131 | CALL CUBRUL(F, VEC, W(1,1), IDATA, RDATA)
| 1
Error: Interface mismatch in dummy procedure ‘f’ at (1): 'f' is not a function
cubtri.f:170:20:
170 | CALL CUBRUL(F, VEC, W(1,J), IDATA, RDATA)
| 1
Error: Interface mismatch in dummy procedure ‘f’ at (1): 'f' is not a function
make: *** [Makefile:11: cubtri.o] Error 1
$ So we are sticking with the binaries from the apt
repository and the default Spooles and internal solvers.